ASKE Month 13 Milestone Report¶
System performance statistics¶
EMMAA currenty manages a total of 11 models. Eight of these models are fully machine-maintained and represent various diseases (7 models) and pathways (1 model). Two models are based on expert-curated natural language, then linked to literature evidence and tested automatically. Finally, one model represents a set of causal factors affecting food insecurity, i.e., is outside the domain of molecular biology.
To quantify the performance of the system in terms of extending and testing/ analyzing models, we plotted the distribution of (1) number of new statements added (2) number of new tests applied and (3) change in the test pass ratio for each of the machine-maintained biology models each day.
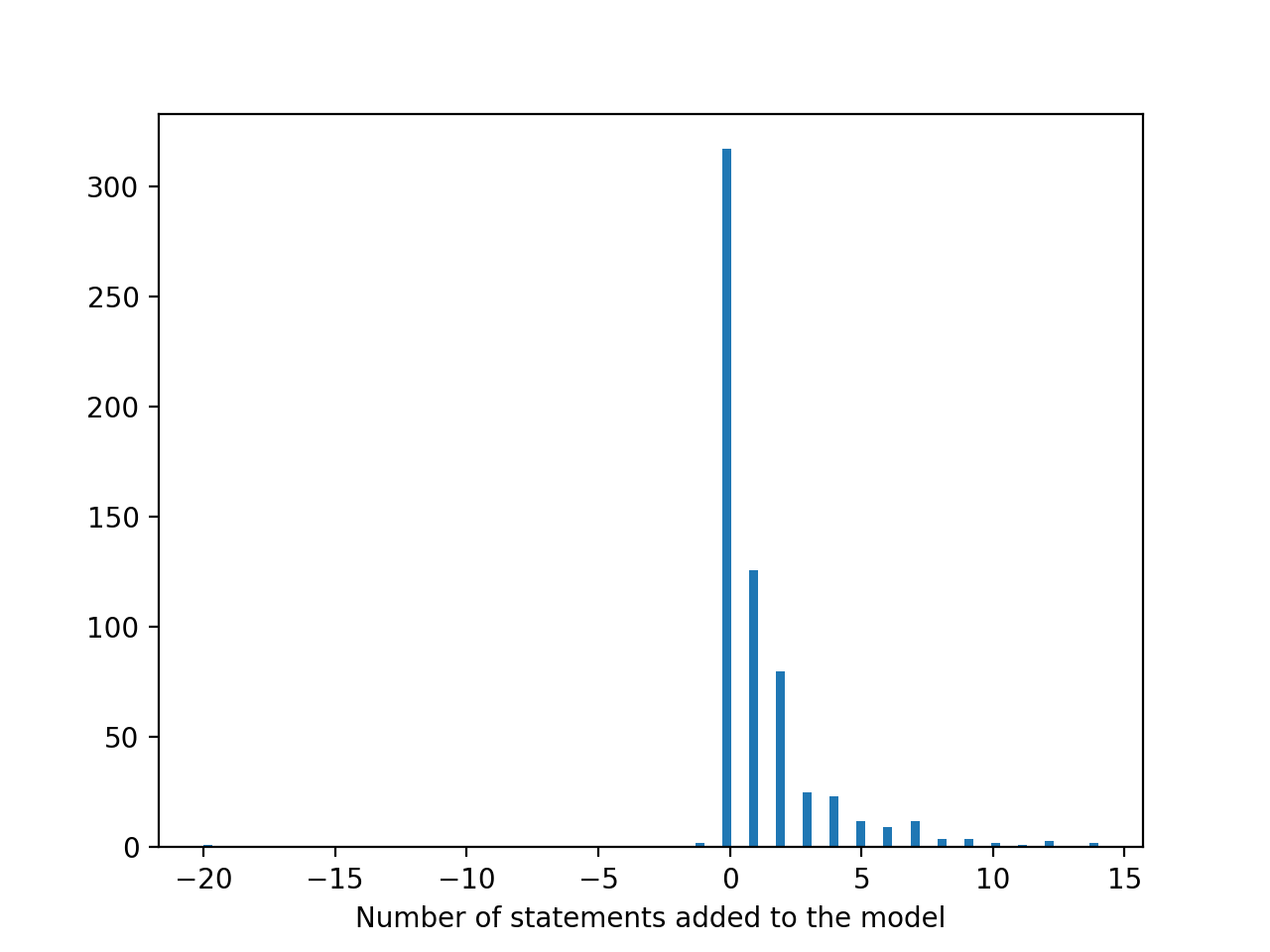
Histogram of the number of new statements added to each model each day. As we can see, the change in the number of statements is often zero (i.e., no new mechanisms were found relevant to the given model), but otherwise is between 1-15 new statements per day. In some cases, the assembly procedure removes previously existing mechanisms from the model, thereby making the number of statements added negative.
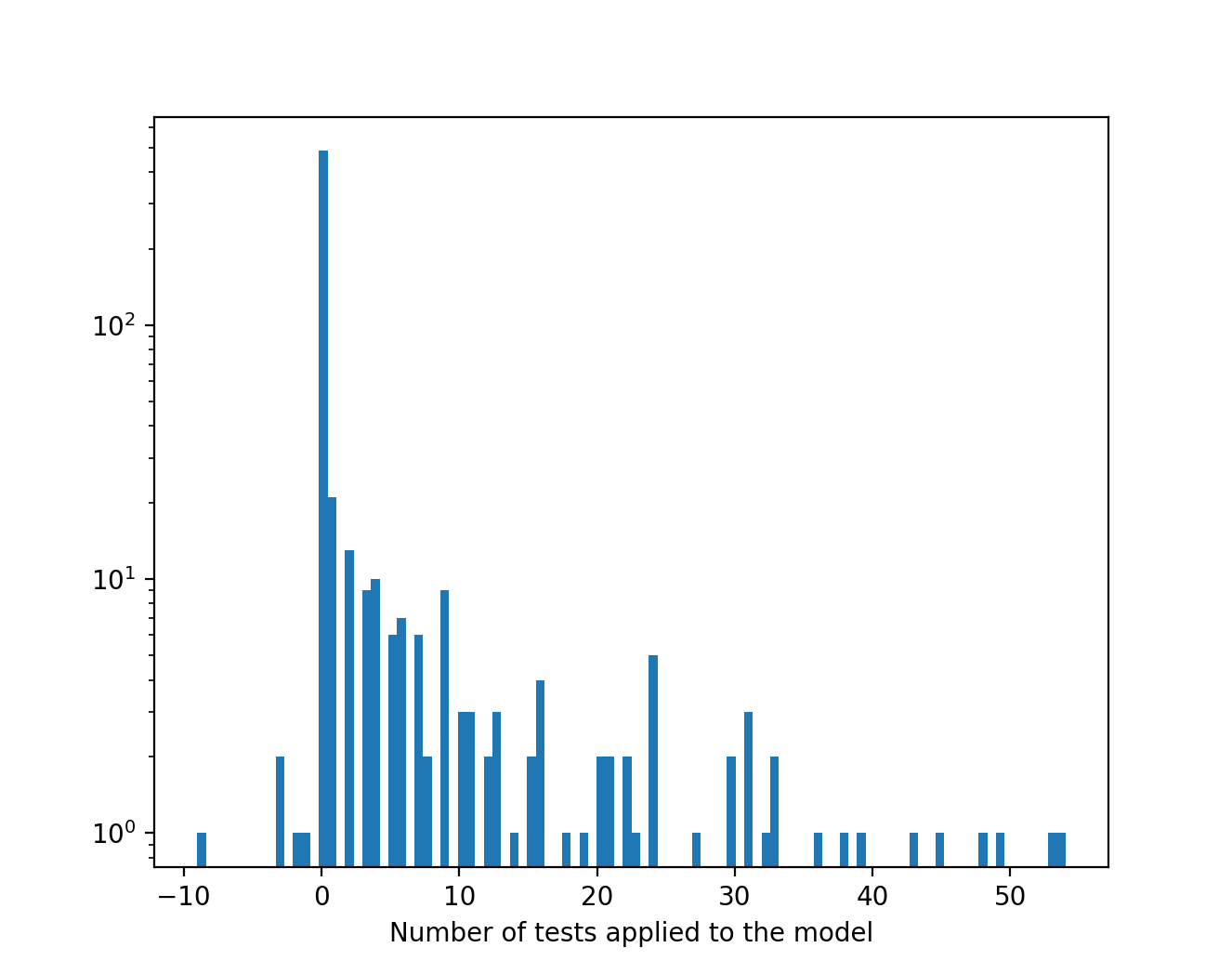
Histogram of the number of new applied tests each day. Typically, if new statements are added to a model, the number of applied tests can increase. As shown in the histogram, new mechanisms added to a model often result in dozens of new test being applicable to the model.
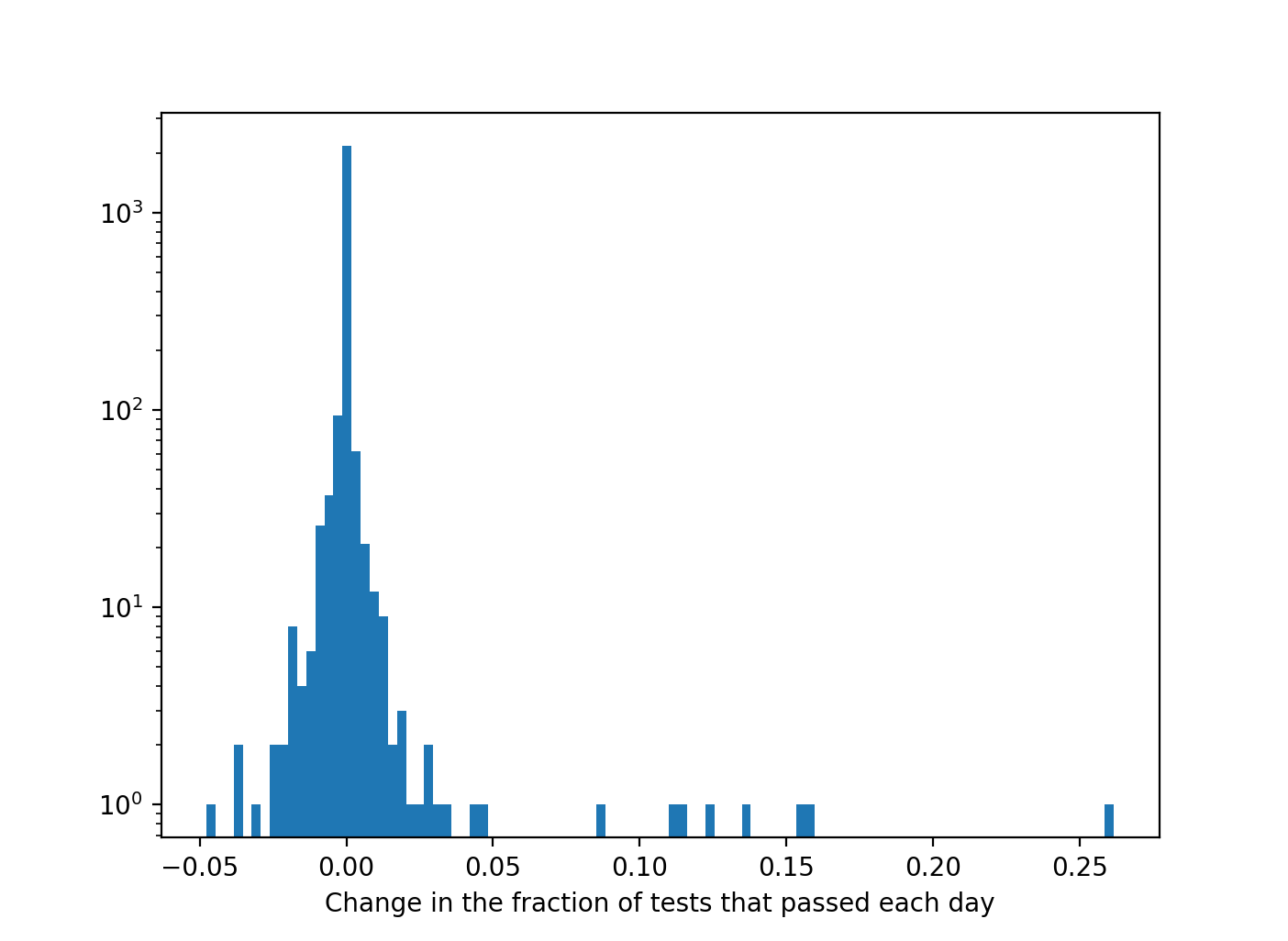
Histogram of the change in the fraction of tests that pass (across all four modeling formalisms, PySB, PyBEL, signed graph, unsigned graph) each day compared to the previous day. While small fractional changes are more common, in some cases, model extensions (or changes to model assembly) resulted in large jumps in test pass ratio of 5-25%.
References¶
Aldridge, B. B., Burke, J. M., Lauffenburger, D. A., & Sorger, P. K. (2006). Physicochemical modelling of cell signalling pathways. Nature cell biology, 8(11), 1195.
Gyori, B. M., Bachman, J. A., Subramanian, K., Muhlich, J. L., Galescu, L., & Sorger, P. K. (2017). From word models to executable models of signaling networks using automated assembly. Molecular systems biology, 13(11).
Ananiadou, S., & McNaught, J. (2005). Text mining for biology and biomedicine (pp. 1-12). London: Artech House.
Valenzuela-Escárcega, M. A., Babur, Ö., Hahn-Powell, G., Bell, D., Hicks, T., Noriega-Atala, E., … & Morrison, C. T. (2018). Large-scale automated machine reading discovers new cancer-driving mechanisms. Database, 2018.
McDonald, D., Friedman, S., Paullada, A., Bobrow, R., & Burstein, M. (2016, March). Extending biology models with deep NLP over scientific articles. In Workshops at the Thirtieth AAAI Conference on Artificial Intelligence.
Allen, J., de Beaumont, W., Galescu, L., & Teng, C. M. (2015, July). Complex Event Extraction using DRUM. In Proceedings of BioNLP 15 (pp. 1-11).
Torii, M., Arighi, C. N., Li, G., Wang, Q., Wu, C. H., & Vijay-Shanker, K. (2015). RLIMS-P 2.0: a generalizable rule-based information extraction system for literature mining of protein phosphorylation information. IEEE/ACM Transactions on Computational Biology and Bioinformatics (TCBB), 12(1), 17-29.
Barbosa, G. C., Wong, Z., Hahn-Powell, G., Bell, D., Sharp, R., Valenzuela-Escárcega, M. A., & Surdeanu, M. (2019, June). Enabling Search and Collaborative Assembly of Causal Interactions Extracted from Multilingual and Multi-domain Free Text. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics (Demonstrations) (pp. 12-17).
Cerami, E. G., Gross, B. E., Demir, E., Rodchenkov, I., Babur, Ö., Anwar, N., … & Sander, C. (2010). Pathway Commons, a web resource for biological pathway data. Nucleic acids research, 39, D685-D690.
Croft, D., Mundo, A. F., Haw, R., Milacic, M., Weiser, J., Wu, G., … & Jassal, B. (2013). The Reactome pathway knowledgebase. Nucleic acids research, 42(D1), D472-D477.
Keenan, A. B., Jenkins, S. L., Jagodnik, K. M., Koplev, S., He, E., Torre, D., … & Kuleshov, M. V. (2018). The library of integrated network-based cellular signatures NIH program: system-level cataloging of human cells response to perturbations. Cell systems, 6(1), 13-24.
Tomczak, K., Czerwińska, P., & Wiznerowicz, M. (2015). The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology, 19(1A), A68.