ASKE Month 7 Milestone Report¶
Repositioning EMMAA within the ASKE framework of modeling layers¶
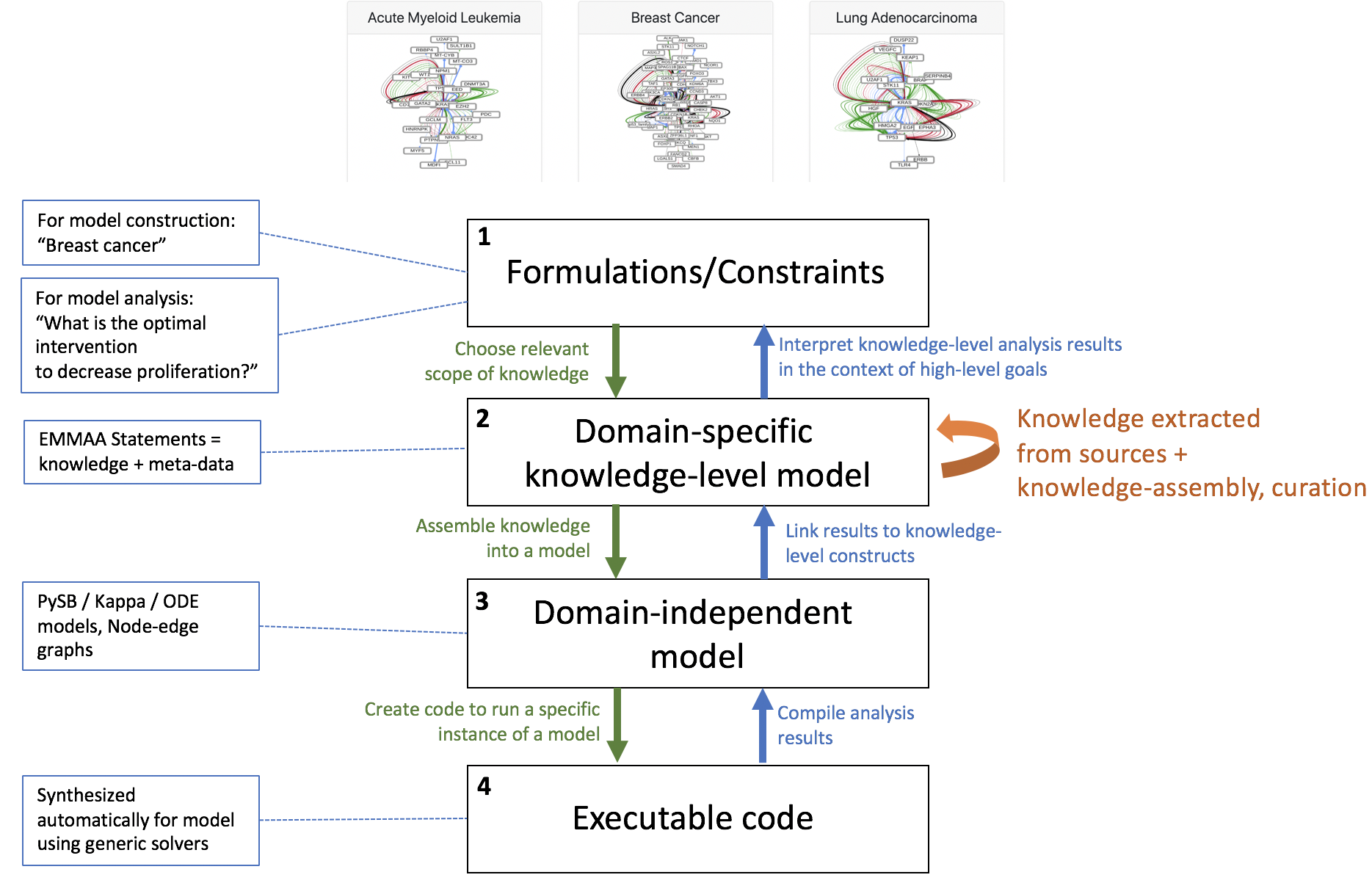
Based on discussions at the ASKE 6-month PI meeting we have been reformulating how our approach in EMMAA relates to the three proposed modeling representation levels. One outcome of the meeting was an emerging consensus that the middle “model” level represents domain-independent model representations that are not yet executable. Examples discussed included linear regression models, polynomial functions, ordinary differential equations, etc. Prior to this discussion we had considered this layer, representing classes of mathematical models, to be the “bottom”; however, we now recognize that a model at this level is not yet executable because it must first be coupled to a particular simulation or inference procedure.
With this in mind, we feel that our approach requires the definition of an additional layer, sitting between the topmost (level 1, “formulations/constraints”) and the mathematical modeling layer (now level 3). This layer corresponds to networks of EMMAA/INDRA Statements: representations of a particular subset of domain knowledge. A knowledge network at this layer may be formulated based on requirements/constraints specified in level 1 (e.g., “a knowledge model of breast cancer”, or “all signaling pathways with a bowtie architecture”). In turn, this knowledge network can be used to generate different analytical/mathematical models at level 3 (boolean networks, rule-based models, analytical/mathematical model ODEs, etc.).
One focus of the discussion during the PI meeting was on the potential for integration of ASKE modeling frameworks via the domain-independent level 3. Integration at this level would allow tools for model analysis, simulation, expansion, etc. to be reused between teams. At this layer domain-specific considerations may still apply but they will have been converted into syntactic constraints expressed in the language of the particular modeling formalism. One example relevant to biology is the formulation of ODE models: while in a domain-independent sense the class of all ordinary differential equation models is quite large, biological models typically make use of a highly restricted subset of mathematical functions. In a “mass-action” reaction model, for example, the right hand side function consists strictly of a linear combination of products of the concentration variables. This (semantic) biochemical constraint could be expressed in the (syntactic) language of mathematical functions to allow the application of tools for model expansion, simulation, etc.
Since the PI meeting we have also concluded that important model inference and transformation procedures can occur at layers other than level 3, and that these operations can occur within, not just across layers. For example, in INDRA there are a set of related procedures that we collectively refer to as “knowledge assembly” or “pre-assembly”: identifying subsumption relationships, inferring and applying belief scores, identifying statement relationships, etc. Both the information considered in these operations, and the operations themselves explicitly make use of domain-specific knowledge, and all take INDRA Statements as input and produce INDRA Statements as output. These steps are referred to as “pre-assembly” to differentiate them from the step of assembly, which denotes the transformation of knowledge-level information (level 2) from model-level information (level 3).
Use cases for the EMMAA system (and ASKE systems in general)¶
Push Science¶
In 2015, Paul Cohen defined “push scholarship” as: “[…] instead of pulling results into our heads, we push results into machine-maintained big mechanisms, where they can be examined by anyone. This could change science profoundly.”
ASKE systems have the potential to go beyond this ambitious goal by:
- actively searching for new discoveries and data,
- autonomously updating a set of models by integrating new discoveries,
- designing model analysis experiments to understand the effect of this new knowledge
- reporting the effect of new discoveries on scientific questions relevant to the user
In other words, novel, relevant implications of discoveries, as soon as they appear, are “pushed” to scientists.
Monitoring reproducibility¶
About 3,600 new publications appear each day on PubMed, in biomedicine alone. Using automated model extension and analysis, ASKE systems can evaluate newly reported mechanisms against experimental observations (data) and vice versa. Reported mechanisms that aren’t supported by prior observations, as well as observations that don’t make sense with respect to existing models can be detected. This technology can help address some aspects of the reproducibility crisis in a principled way.
Automated scientific discovery¶
There is a large body of unexplained observations (i.e., open scientific questions for which no underlying mechanistic explanation is known) appearing in the biomedical literature and in data stores An ASKE system that immediately aggregates and models new knowledge and evaluates its implications with respect to unexplained observations, is likely to be the first to notice that a previously unexplained observation can now be explained. Novel candidate explanations to observations constructed automatically using ASKE systems can be experimentally confirmed and published.