ASKE-E Month 4 Milestone Report¶
EMMAA Neurofibromatosis Models and NF Hackathon Prize¶
During this reporting period we won one of the top prizes in the “Hack for NF”, a six-week event sponsored by the Children’s Tumor Foundation to develop novel software relevant to neurofibromatosis (NF), a set of cancer syndromes that affect children.
Our submission consisted of two causal models of NF deployed in EMMAA. The first model was built directly from text mining the 18,000 PubMed articles about NF; it contains approximately 9,000 statements about the functions and interactions of NF1, NF2, and other entities mentioned in those articles. Unlike the other cancer-related models in EMMAA, the NF model does not specify an explicit list of disease-relevant proteins: the scope of the model is defined strictly by neurofibromatosis keyword search terms. This keeps the content of the model as disease-specific as possible, with the model serving as a comprehensive representation of what is known about NF.
For the second part of our submission, we substantially expanded our curated Ras signaling model to include mechanisms relevant to NF1 and NF2 signaling. The model is transparent even for non-modelers because it is built from ~200 declarative English sentences and automaticalaly assembled by INDRA. In an iterative, test-driven process, we used the reported causal relationships from the literature-based NF model that were unexplainable by the curated model to both 1) identify errors in the literature derived model and 2) discover necessary extensions to the curated model.
As an example, the literature-based model contains the observation that NF2 inhibits PAK1. The extended curated model shows that this this finding can be explained by a mechanistic path whereby NF2 competes Angiomotin (AMOT) away from inhibiting ARHGAP17, allowing ARHGAP17 to inhibit CDC42, which would otherwise activate PAK1.
As a further demonstration of the scientific value of automated model analysis, we converted drug screening data from NF1 and NF2 cell lines into EMMAA tests and checked the literature-derived model against them. Interestingly, we found that while the causal paths identified by the models were typically short, involving paths with a single intermediate node (i.e., drug->protein->cell proliferation) the explanatory nodes were highly context-specific, in some cases having been previously identified in the literature as therapeutic vulnerabilities for NF cell lines.
We see the two types of models (curated and literature-derived) as working synergistically to explain experimental results and accumulate actionable knowledge, as shown in the diagram below.
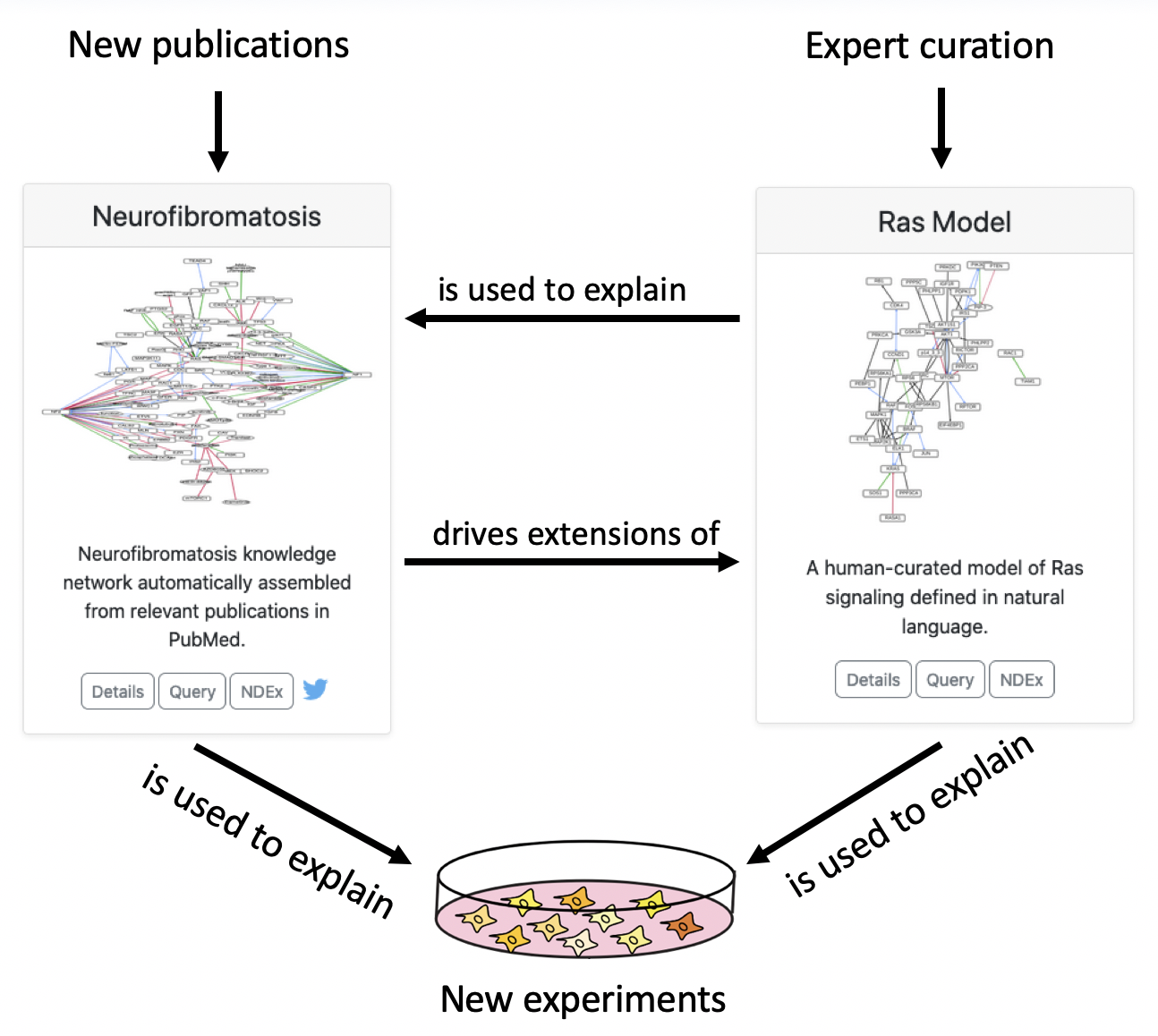
For this hackathon entry, we won one of three top prizes. The press release from the Children’s Tumor Foundation can be found here, and a video presentation describing our project can be found here.
Rapid initialization of EMMAA models from literature for two new diseases¶
The new Literature Prior module module makes the instantion of EMMAA models based on a subset of the scientific literature straightforward. As input, the class takes a list of PubMed search terms and optionally a list of Medical Subject Headings. It then automatically identifies relevant publications, and collects all statements from text mining that were extracted from these papers. The model is then uploaded to AWS and is available for daily updates and access via the dashboard. We used this method to start two new EMMAA models, for vitiligo and multiple sclerosis.
Downloading EMMAA models in alternative formats¶
The knowledge assembly approach in EMMAA allows exporting each model in multiple different modeling formalisms. In fact, EMMAA internally uses four different modeling formalisms (PySB, PyBEL, signed graph and unsigned graph) for querying and analysis. However, these formats, and other community standards have not been made available to users through the EMMAA dashboard.
We added multiple exports for each model that are generated during each model update (typically daily) and are available through the EMMAA dashboard. Each model has the following export formats available:
- json.gz: A gzipped INDRA Statement JSON dump.
- jsonl: An uncompressed dump of INDRA Statement JSONs with one statement per line.
- indranet: A tabular (tsv) file where each row represents a single binary interaction between two entities. This format is ideal for building networks from an EMMAA model.
Models that support PyBEL analysis provide a pybel export. In addition, models that support analysis at the rule-based executable level are exported into the following formats:
- bngl: BioNetGen model representation (http://bionetgen.org/)
- kappa: Kappa model representation (https://kappalanguage.org/)
Finally, models that support reaction-network based analysis are exported into these formats:
- sbml: Systems Biology Markup Language (http://sbml.org/)
- sbgn: Systems Biology Graphical Notation (https://sbgn.github.io/)